Towards End-to-End Knowledge Discovery in Complex Brain Networks
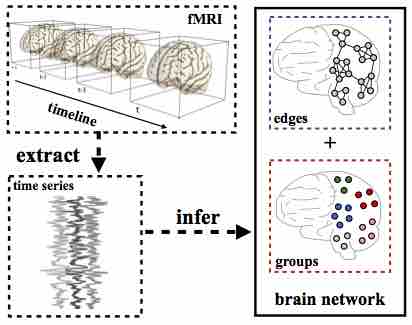
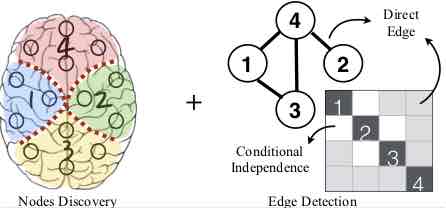
Modeling the brain as a networked system has become one of the most pervasive paradigms in neuroscience. However, knowledge discovery on brain networks is still extremely hard. Researchers must be taught in tedious detail how to design the pipeline of data processing, tune the parameters in different processing stages (e.g., node discovery and edge discovery), and must constantly be adjusted for changes in any stage of the pipeline (from data collection to modeling). The laborious teaching and adjustment incur huge expenses, and pose a key bottleneck for the widespread adoption of brain network analysis in clinical and research studies. There is a significant gap between the demand of neuroscience researchers and existing knowledge discovery systems for brain networks. To bridge this gap, we need to go beyond the pipeline-based approaches, where the multiple stages of the knowledge discovery are studied in isolation. In this project, we aim to design end-to-end methods for knowledge discovery in brain networks. Different stages of the knowledge discovery on brain networks are integrated and studied together. These stages include: (1) node discovery: also called "brain parcellation"; (2) edge discovery: also known as "connectome" in neuroscience; (3) feature mining: which aims to extract subgraph features from the networks; (4) supervised learning: building statistical learning models on the subgraph features.Acknowledgements:
This project is gnerously supported by NSF Grant #1718310.Papers
-
Collective Discovery of Brain Networks with Unknown Groups
International Joint Conference on Neural Networks (IJCNN), 2017
-
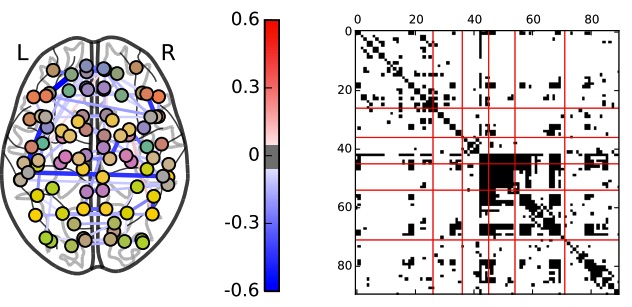
Unified and Contrasting Graphical Lasso for Brain Network Discovery
SIAM International Conference on Data Mining (SDM), 2017
-
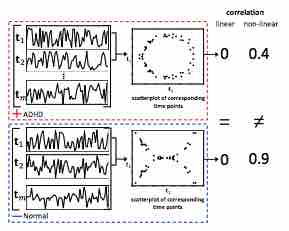
Identifying Deep Contrasting Networks from Time Series Data: Application to Brain Network Analysis
SIAM International Conference on Data Mining (SDM), 2017
-
Coherent Graphical Lasso for Brain Network Discovery
IEEE International Conference on Data Mining (ICDM), 2018
-
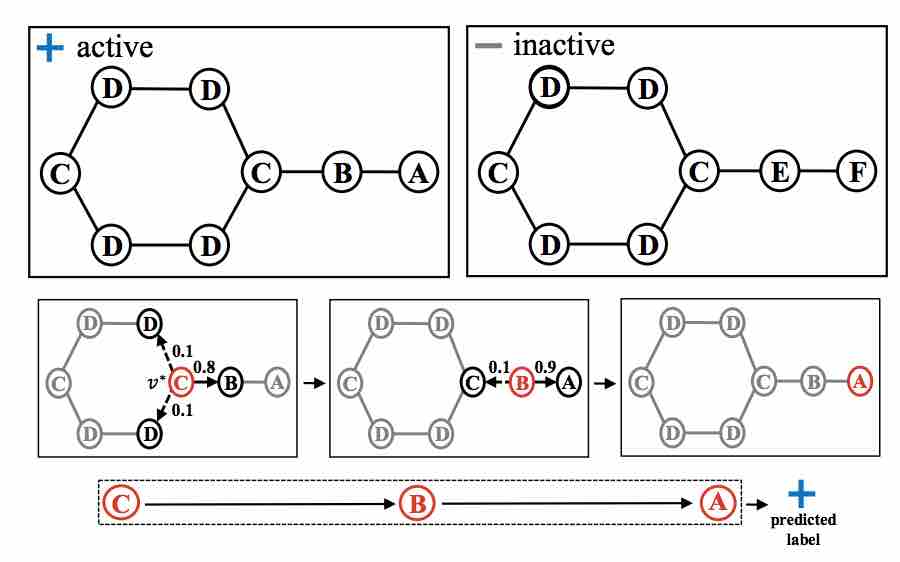
Graph Classification using Structural Attention
ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD), 2018
-
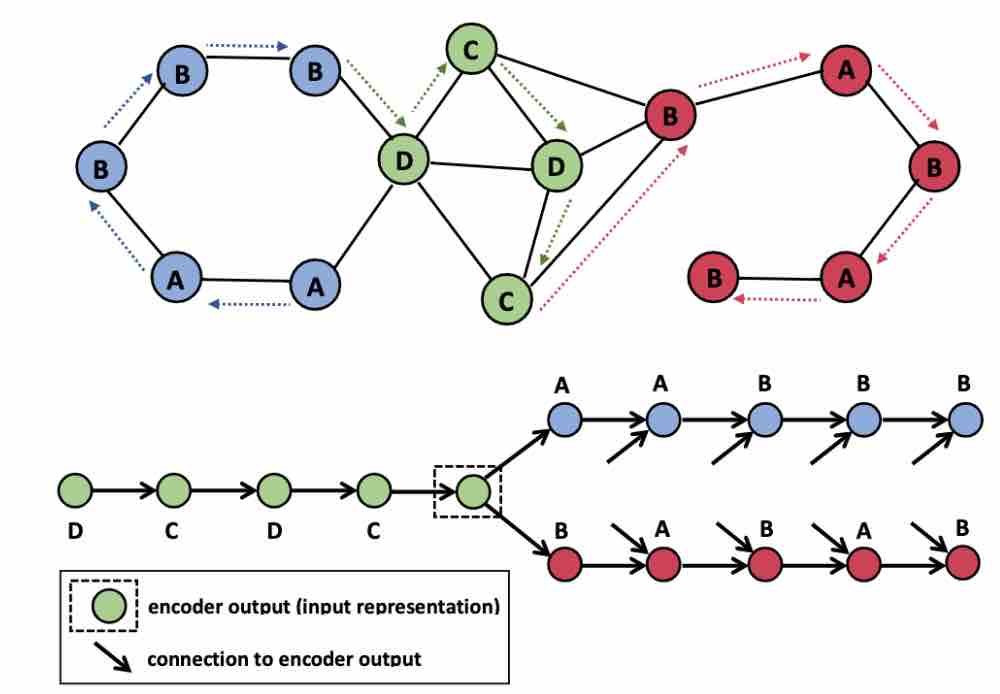
Learning Compact Graph Representations via an Encoder-Decoder Network
Applied Network Science (APNS), 2019
-
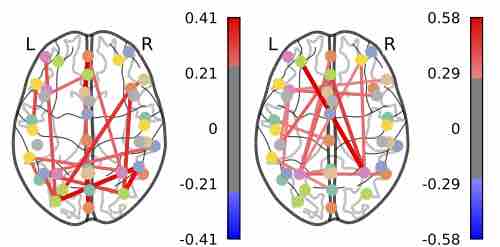
Gaussian Mixture Graphical Lasso with Application to Edge Detection in Brain Networks
IEEE International Conference on Big Data (BIGDATA), 2020
-
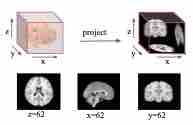
Dual-Attention Recurrent Networks for Affine Registration of Neuroimaging Data
SIAM International Conference on Data Mining (SDM), 2020
-
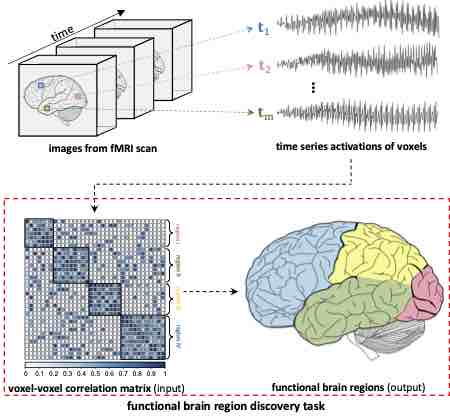
Deep Parametric Model for Discovering Group-cohesive Functional Brain Regions
SIAM International Conference on Data Mining (SDM), 2020
-
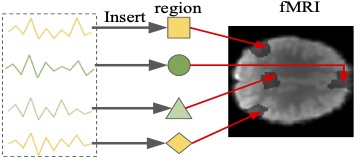
Recurrent Networks for Guided Multi-Attention Classification
ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD), 2020





