Introduction
Foldit is a puzzle game where the player manipulates the structure of a protein in 3D space to reach as high a score as possible. The protein structure and properties are accurate to those of an actual protein in the real world. The game records the algorithms that players use to get their high scores and uses the same algorithms to steer scientific research in finding better solutions to real life problems.
Below is a detailed analysis of this game roughly following Brian Winn's1 Design/Play/Experience framework, including:
Learning
What’s really interesting about Foldit is that both players and scientists are learning from the gameplay. The data collected from the puzzles are compared to the data produced by powerful machines running protein folding simulations to see if a person’s problem solving intuition is more time efficient than the algorithms that computers are currently using.
The players working through the puzzles are taught:- The difference between hydrophilic and hydrophobic amino acids and their roles and properties in protein folding.
- How hydrogen bonds are used to solidly hold the structure of a protein.
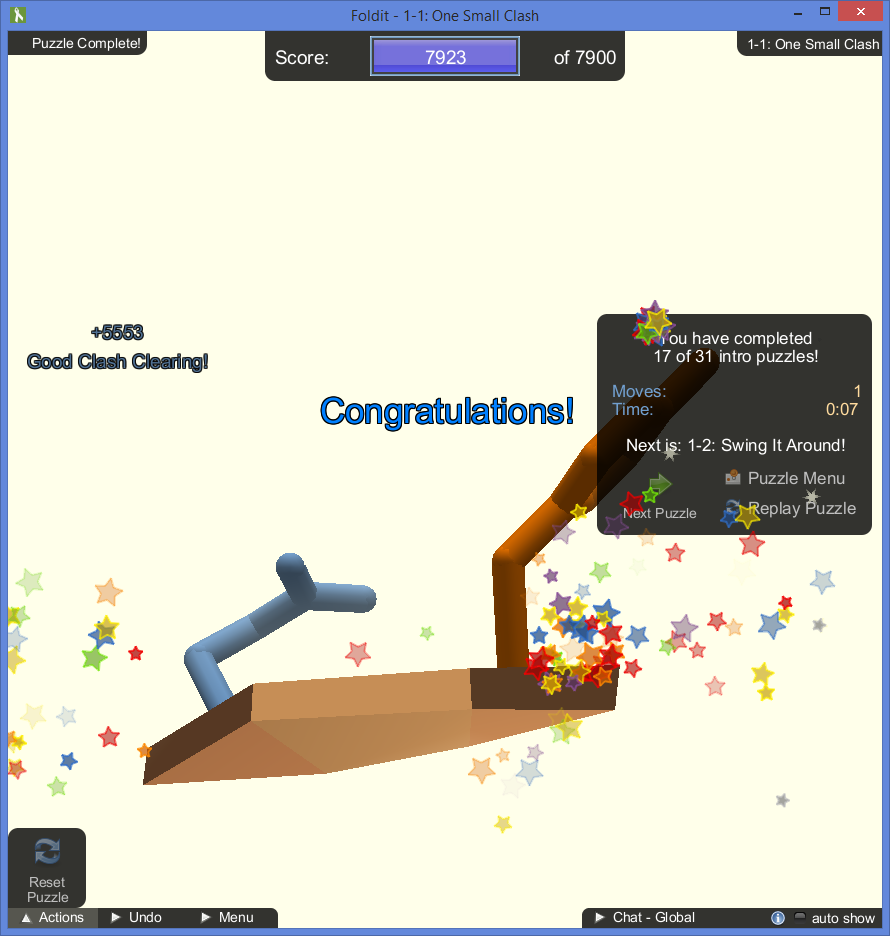
- Amino acid sidechains cannot occupy the same space or they will “clash”.
- Gaps in the center of a protein are inefficient and can filled more tightly.
Biological research is further developed through the game by:
- Searching for proteins that more efficiently break apart plant material into biofuel.
- Learn more about the proteins responsible for HIV/AIDS, Cancer and Alzheimer's Disease.
- Create new proteins that could potentially make exciting breakthroughs, such as cleaning carbon dioxide out of the atmosphere.
Storytelling
The game has no story, setting, characters, or narratives. Instead, it gives the players proteins with puzzle challenges to achieve the highest possible scores with a set of tools given. We do not think the game interferes with the learning process, since the players engage themselves to solve the challenges through learning and utilizing the processes of folding available.

Gameplay
There are three main goals of Foldit:
- Protein structure prediction.
- Take folding strategies from human players, and automate them.
- Protein designing.
The first goal is to have human players work on proteins that do not have a known structure, but it requires attracting the attention of scientists and biotech companies and convincing them that this process is effective.
The second goal is to take folding strategies that human players have come up with, and automate these strategies to improve protein-prediction software.
The third goal is to design new proteins. A protein designer is basically an engineer who uses proteins to tackle problems such as disabling a virus, or absorbing carbon dioxide from the atmosphere.
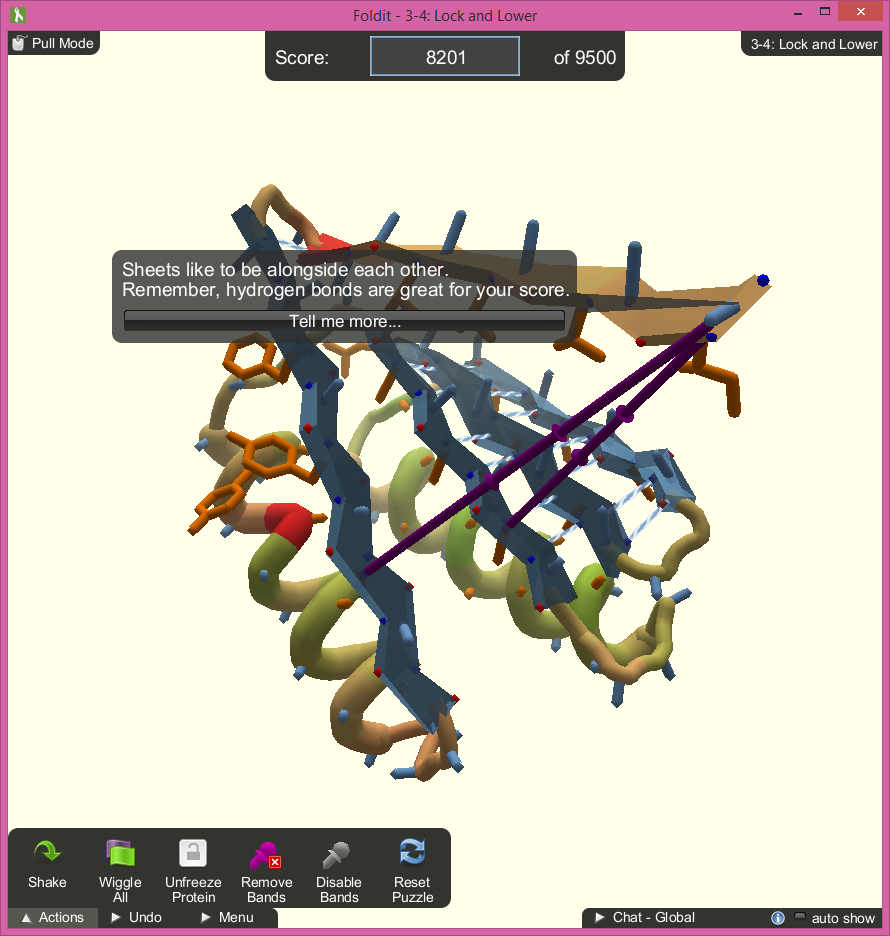
The game allows the player to fold, which is by clicking and dragging the protein’s amino acids to make it face a certain direction. The game also allows the player to rotate the protein around, so the player can see the proteins at all angles. Folding the protein structure, such as hiding the hydrophobics and exposing the hydrophillics, will increase the score count. The player can also create hydrogen bonds by dragging sheets close together, which is the best way of scoring points in the game. As the player folds, the game tallies the score, which tells the player how successful the proteins are folded.
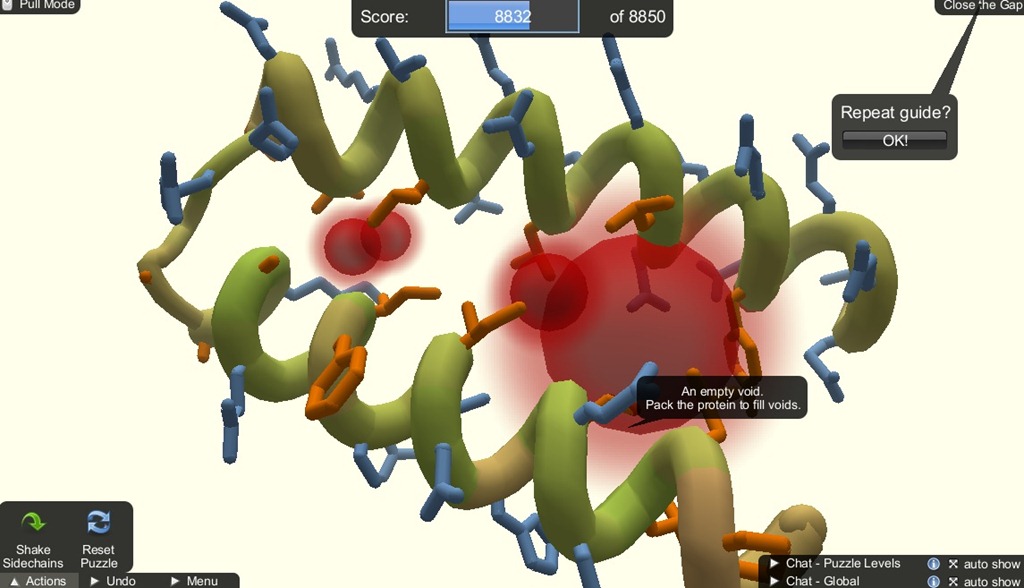
The shaking mechanic is for setting the hydrophobic and hydrophillic sidechains to its optimal state by pulling the sidechains. If there are sidechain clashes, shaking will cause them to face away from each other.
The wiggle mechanic causes the structure of the protein to move around, gaining more points if possible, but never decreasing the player’s score. Wiggling gives players an option for more precisely and automatically moving the protein into its intended position without accidentally dragging it too far out of place. It also provides players an easy way to make progress without tediously moving parts of the protein an inch at a time.
Rubber bands give players the power of pulling sections of the protein together without manually moving each piece. Combined with the wiggle ability, rubber bands give the wiggling a focused direction to maximize the points the player will earn from it.
The player can also lock a section of the protein in place so that it won’t be affected by wiggling. Once the player has made some high-value hydrogen bonds, they may want to lock the bonds in place so that they don’t accidentally drag or rotate them apart .
While wiggling, shaking and rotating the pieces into place will earn the player more points as they are fiddled with, the rebuild mechanic is more directly educational and is required to solve some of the puzzles. Certain sections of the protein can have their shape changed to score more points. In the tutorials, there is an intended way for the proteins to be rebuilt, but in later puzzles the player is free to rebuild any section they think is inefficient.
Once the player’s score exceeds the target score in the tutorial, the player will beat the level and move on to the next tutorial. In the actual puzzles and levels designed by other players, players will compete for the high score on a leaderboard. Some of the puzzles don’t have a known solution, so players will try to get the highest maximum score.
User Experience
The interface is very simple, but does its job well. The screen is mostly kept clear of HUD elements. The actions that a player can perform are tucked away in a small menu in one corner and the player’s current score and goal score are displayed at the top. There is also a global chat feature that can be used in the bottom right corner, but is minimized when not in use. The minimal HUD interface is appreciated as the model itself takes up a large portion of the display and has many small details that need to be focused on. The player needs as much space as possible to rotate and manipulate the pieces of the protein.
During the tutorials, tips will pop up in speech bubbles in the center of the screen. The bubbles point directly to the spot on the protein that the tip is referring to, even as the player rotates the model. This does a good job or directing the player’s focus and helps explain the complicated mechanics well. The popups can get annoying after resetting the same level multiple times, but they only appear on the tutorials and can be exited.
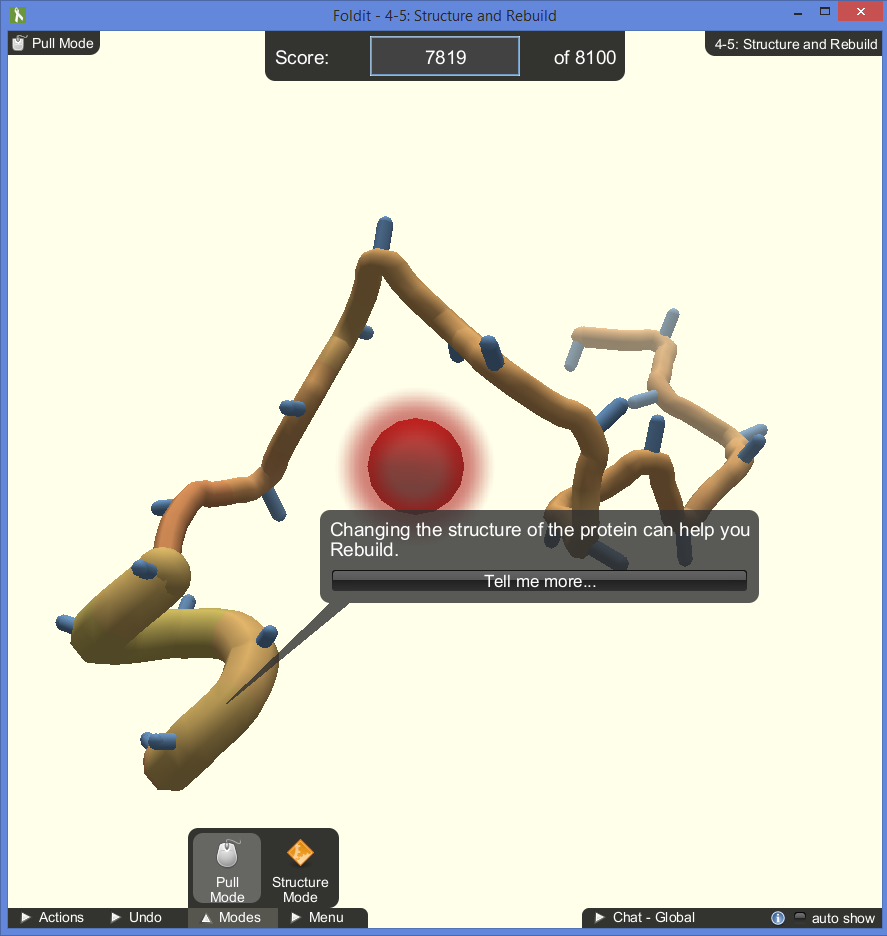
The game does a nice job of identifying what sections of the protein need tending to by spawning symbols directly on the model:
- Red bubbles identify gaps in the structure that need to be closed.
- Yellow bubbles identify hydrophobic sidechains that are exposed and should be hidden away inside the protein.
- Red spikes identify clashes in sidechains that need to be pulled apart more.
- Blue and white striped lines show hydrogen bonds that greatly increase the player’s score
- Sections of the protein base structure turn red when in trouble and green when they are in a good position.
By placing these indicators on the model, the player knows exactly what spots to fix and give a quick way of noticing whether a change helped increase the score or made everything worse.
When a change makes a negative impact on the structure, a red number will show how many points were lost. When a good change is made, a blue number appears showing the point gain. This adds a nice sense of satisfaction as the player makes small tweaks to the structure to try to scrape together as many points as possible. There is a big feeling of accomplishment when a large blue number appears as everything clicks together.
The controls are fairly standard. Anyone who is familiar with 3D modelling software will immediately understand how it works. New players who have no experience working in 3D could have trouble figuring out how to get the best viewing angle. One problem we noticed is that left clicking and dragging is how the player rotates the camera when the mouse is not overlapping the protein, but is also how parts of the protein are repositioned. There were quite a few times where we would attempt to rotate the camera but accidentally mess up the protein instead. Thankfully the game allows the player to undo their actions, making the risks minimal.
Technology
It attempts to apply the human brain’s natural three-dimensional pattern matching and spatial reasoning abilities by making the game in 3D.
The developers used the concept of gamification to design a program that is more appealing and engaging to the public audience, rather than building a useful science tool. This helps to attract more people to do protein folding, especially for those who do not have a scientific education or background.
The developers added the ability to chat, create and join groups, and share puzzle solutions in the game. This is done to encourage players to come up with better puzzle solutions, and in turn, help to solve problems of protein structure prediction.
Assessment
Seeing as how Foldit is used to find better solutions to real world problems, the scientific breakthroughs resulting from the gameplay show how successful the game is. One example of this success is when the developers challenged players to remodel the backbone of a an enzyme to make it catalyze more efficiently [2]. Advanced players were given proteins to restructure as a puzzle in the game and the best solutions in each wave were saved and given back to the players as the next wave. This meant that players were building off of each others’ success and were able to crowdsource the most efficient enzyme.
Because the game provides an in-game chat and a forum for discussion among players, the algorithms or “recipes” that players produce to achieve high scores are shared and iterated upon to continuously improve [4]. Players were able to collaboratively produce algorithms of equivalent quality to algorithms produced by professional scientists in the same amount of time. Even without help from the game developers, players were able to make breakthroughs just by working among themselves.
No assessment could be found for how much a new player who is unfamiliar with biology and protein folding would learn from playing the game. There are activities that have been used in classrooms to help students studying protein folding in school to understand how protein structure is based on amino acid sequences and the bonds that form by twisting wires [3]. To assess Foldit, a sample of students could be presented with a pretest and posttest on the topic where one group of students has played Foldit, another has performed the in-class activity of folding wires and a control group has listened to a typical lecture. The understanding of the material on the test would show if the computer simulation was a better teaching tool than the arts and crafts demo.
Conclusion
We feel the game very much succeeds in its attempts to use crowdsourcing as a way to improve and collect data on protein folding and protein engineering. The game allows players and scientists to explore more drastic changes to the protein than were possible using standard methods, such as directed evolution, in which a large pool of randomly mutated enzymes is screened for mutants that improve the original. One notable example is the creation of an enzyme with more than 18-fold higher activity than the original [6].
However, we feel the game is lacking thorough explanation of protein folding. It may be useful if the explanation from the Foldit FAQs can be seen from inside the game, as well as provide labels for the in-game amino acids the player can interact with. Overall, we deem this game successful.
References
- Winn, Brian. The Design, Play and Experience Framework. In R. Ferdig (Ed.), Handbook of Research on Effective Electronic Gaming in Education. Hershey, PA: IGI Global, 2009, pp. 388-401.
- Christopher B Eiben, Justin B Siegel, Jacob B Bale, Seth Cooper, Firas Khatib, Betty W Shen, Foldit Players, Barry L Stoddard, Zoran Popović, David Baker (2012). Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nature Biotechnology, 2012.
- White, Brian. “A Simple and Effective Protein Folding Activity Suitable for Large Lectures.” Ed. Graham Walker. CBE— Life Sciences Education 5.3 (2006): 264–269. PMC. Web. 12 Feb. 2015.
- Firas Khatib, Seth Cooper, Michael D. Tyka, Kefan Xu, Ilya Makedon, David Baker, Foldit players (2011). Algorithm discovery by protein folding game players. Proceedings of the National Academy of Sciences of the United States of America vol. 108 no. 47 18949-18953, 2011.
- Jessica Marshall. “Online Gamers Achieve First Crowd-Sourced Redesign of Protein” Nature. Web. Jan. 22, 2015. http://www.scientificamerican.com/article/victory-for-crowdsourced-biomolecule2/