[WPI] [CS Dept] [ECE Dept] [cs545/ee545] [Syllabus]
Complete by Class 12
Tomographic Reconstruction
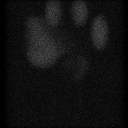
File test_data contains a set of tomographic projections (supplied courtesy of Prof. Michael. A. King, U. Massachusetts Medical Center), the same ones that were used to generate the dataset shown in class. This is simulated Single Photon Emission Computed Tomography (SPECT) data, and incorporates blur, attenuation, and photon statistics (Poisson random variables) such as would be found in real patient data. The simulation used a human body, and shows uptake of the radionuclide by the liver, kidney, ovaries, and heart. Unfortunately, the data is organized feet-end first and head-end last, so that it appears upside down in the following views.
The dataset is organized as 128 columns x 128 rows x 120 projections, where each of the 120 projections is a complete 128 x 128 image. Each datum is a 4 byte floating point number, following the Digital Equipment Corporation byte-ordering convention. That means that reading this dataset on another vendor's machine will not work. Not to worry ...
Program f2di.c was used to convert this large (almost 8 megs) dataset into something a bit more manageable by writing unsigned chars in place of floats. The resulting data file test_data.dat is 1/4 the size of the original. The new data file also has header information attached to it. Note that the header differs slightly from the header format used in previous programs because it needs to handle 2 or 3 dimensional data. Look at the source code to see the header format.
Here is a typical 128 x 128 projection image at projection number 1.

From the 120 projections, 24 were extracted (numbers 1, 6, 11, ...) and made into an animated gif file using a webware program located at http://www.vrl.com/Imaging/animate.html. Most browsers should be capable of displaying this movie -- it is in GIF89 format. It is recommended that you load and view the movie (262 Kbytes).
The GIF files that comprise the movie (converted from TIFFs) can be viewed individually in the image directory. Relate programs may be viewed in the software directory.
As discussed in class, a single point in (x,y) coordinates maps into a
sinusoid in projection coordinates (s, theta).
Program
slice_at_row.c
was used to extract a single row of data.
The result is a 2-D dataset, indexed by column (that is, s) and view
number (that is, theta).
Here is an example, showing row 40.
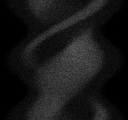
Note that although a full 360 degrees are shown, at 3 degree
intervals,
the data are not quite
symmetric; the sinusoidal patterns are not exact.
This is because this dataset incorporates attentuation and blur, both
of which depend on distance to the detector.
Hence, views taken 180 degrees apart experience differing attenutation
and blur.
This might lead to some errors, which we will ignore, in the reconstruction.
Write a program to reconstruct a slice. You should follow these steps:
![]()
 WPI Home Page |
[CS Dept] [ECE Dept] [cs545/ee545] [Syllabus] |